Welcome to the lab!
We are interested in the ecological and evolutionary factors shaping bacteriophage genomes, and engineering phage genomes as tools for molecular biology.
We use laboratory work and bioinformatics to understand how different genomic parts of the virus affect the virus as a whole, their bacterial hosts, and the microbiomes they exist in.
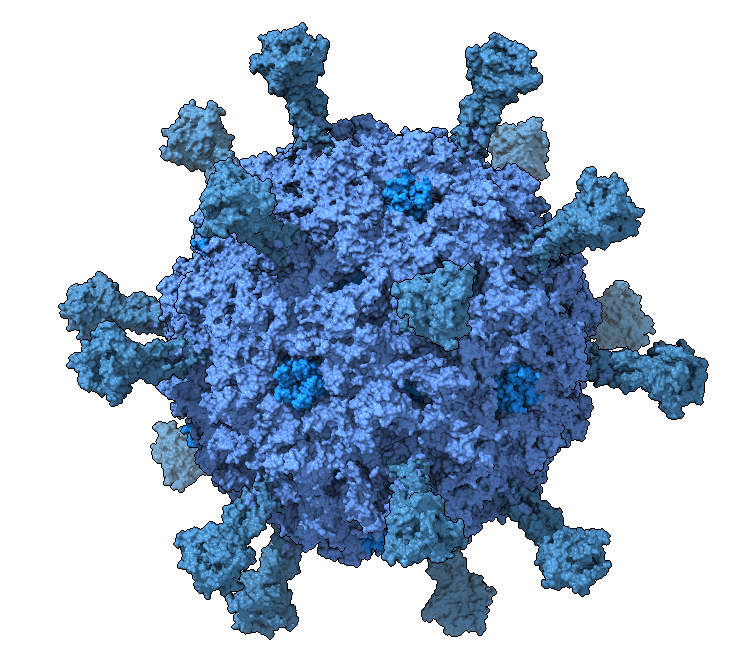
We work by analyzing genomes in silico, synthesizing them in vitro, and generally dissecting and reassembling phages - specifically single-stranded DNA phages of the family Microviridae.
Currently, our focus lies on studying the interaction of microviruses with bacterial hosts and competition with other phages, as well as exploring microvirus diversity in humans and elsewhere.
In the future, we hope to leverage knowledge gained from this research into building customized phages for phage therapy and ecosystem engineering.
News
News
-
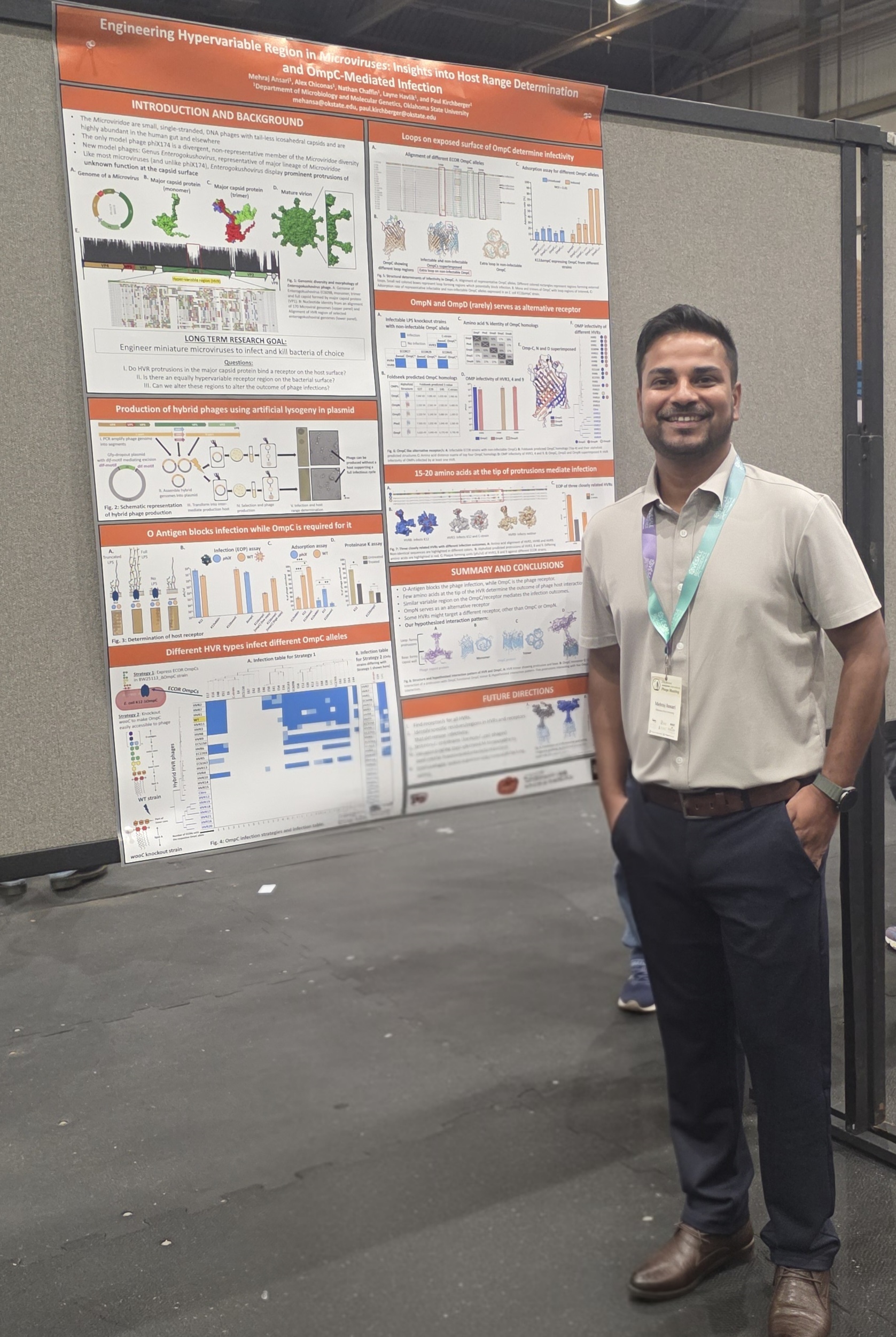
Mehraj wins poster prize at Evergreen Phage Meeting in Knoxville! And Paul presents the newest research on mysterious marine mobile genetic elements
-
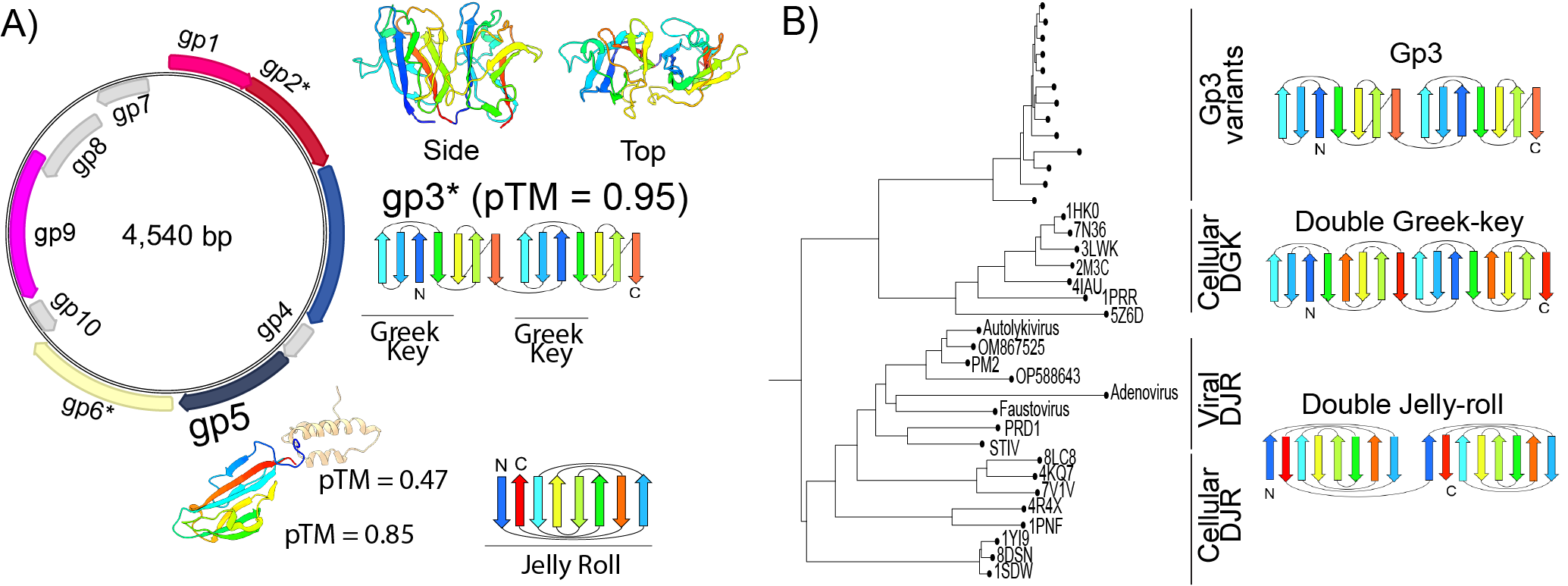
A mysterious new virus? - Preprint! A new preprint in collaboration with Toni Luque from U. Miami describes our accidental discovery of a novel genetic element that looks suspiciously like a virus but does not resemble anything else...
-
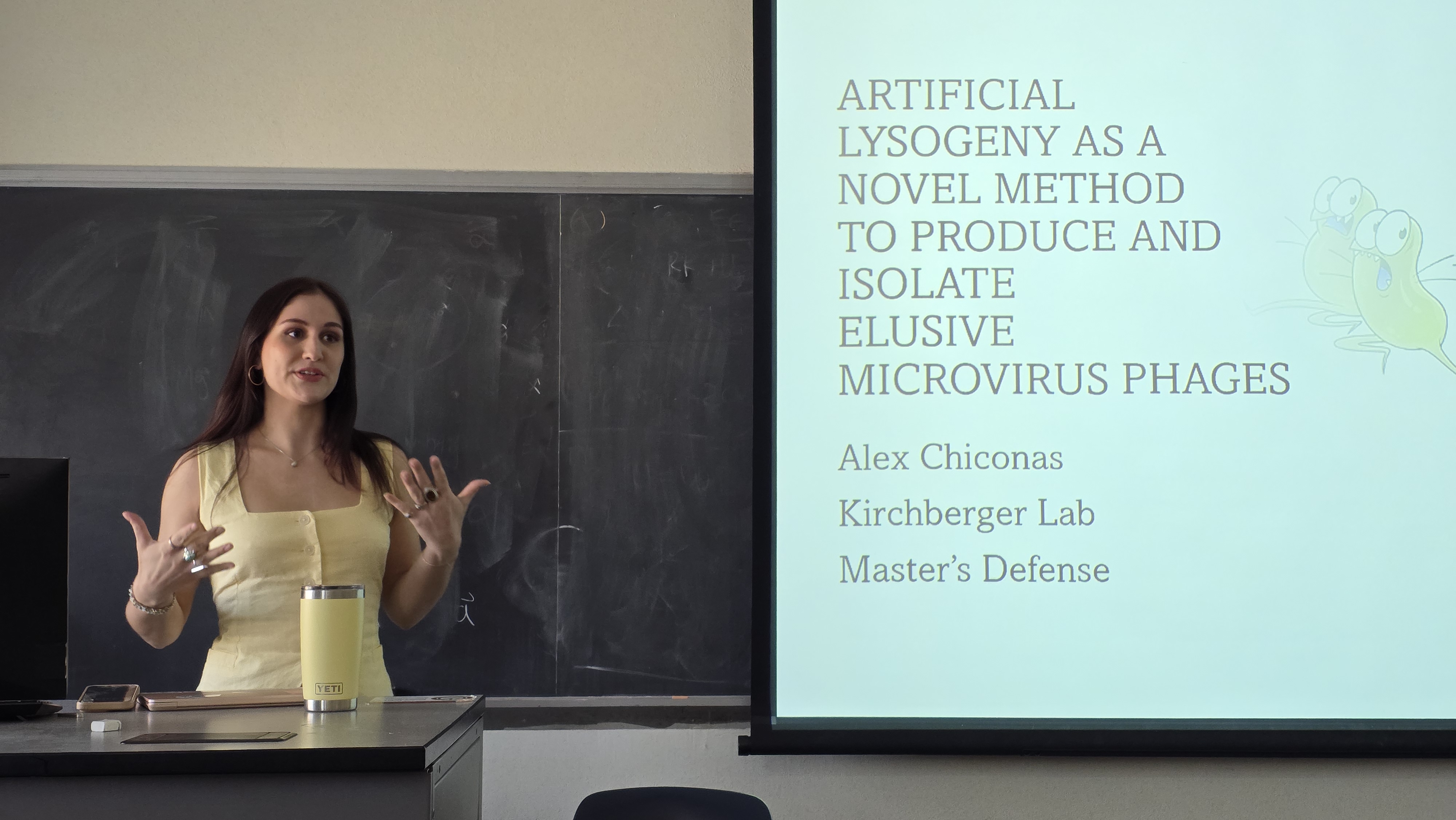
Alex defends her thesis, and Padam gets a job at UNC Chapel Hill! Big News! Alex becomes the first graduate of the Kirchberger Lab - congratulations Master Chiconas! Also, Padam moves on from his short stint in the Kirchberger lab to start a postdoc at UNC Chapel...
-
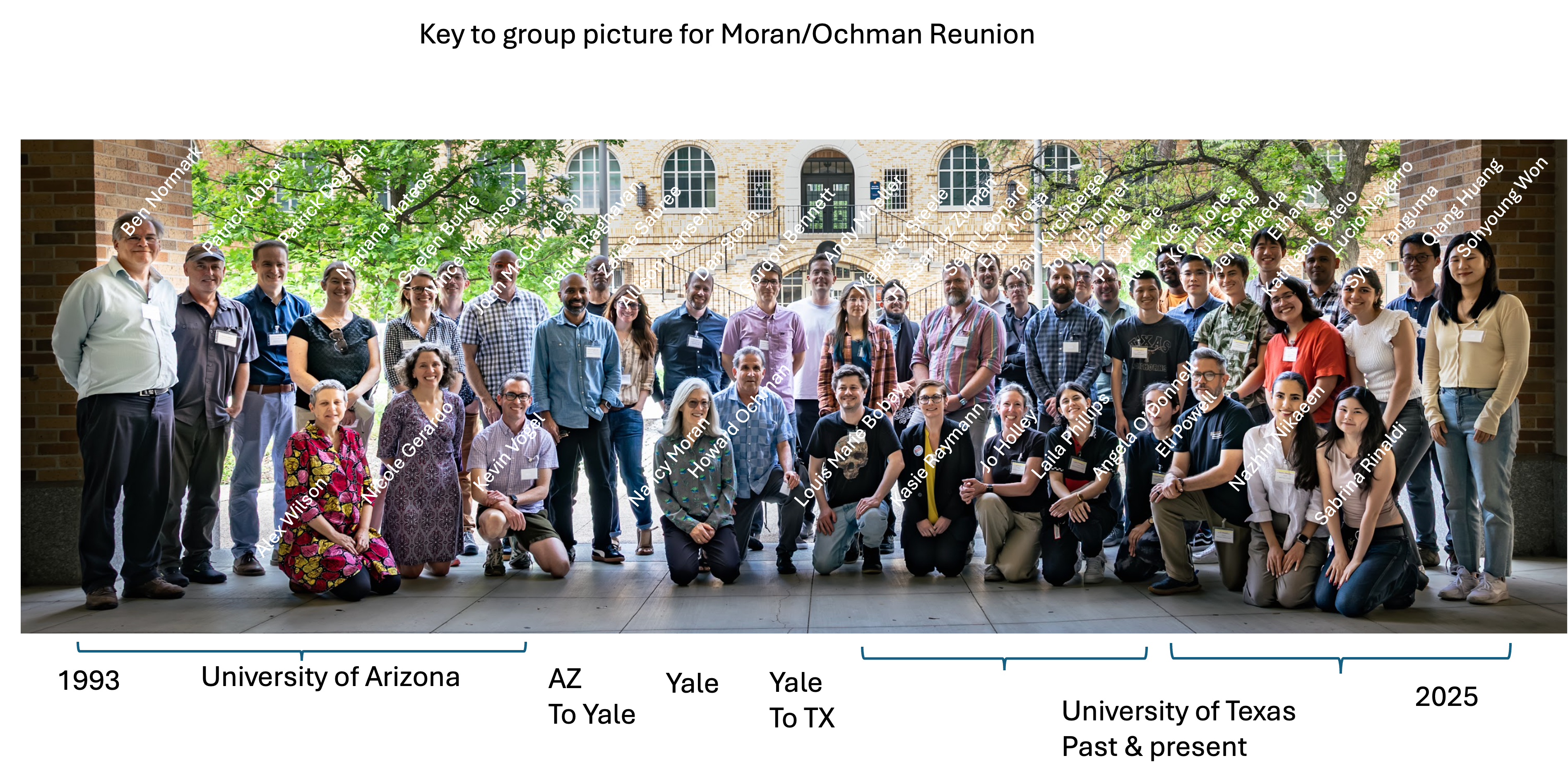
Paul presents at Evolution of Symbiosis, Insects and Microbes Symposium at UT Austin A.K.A. the Howard Ochman & Nancy Moran Lab alumni reunion in lovely Austin Texas.









